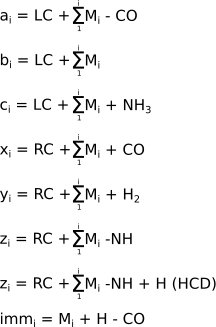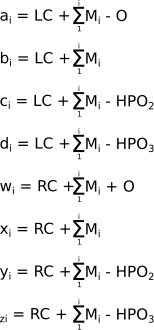massXpert2 User Manual
2 Basics in Polymer Chemistry
This chapter will introduce the basics of polymer chemistry. The way this topic is going to be covered is admittedly biased towards mass spectrometry and biological polymers. Moreover, the aim of this chapter is to provide the reader with the specialized words that will later be used to describe and explain the (inner) workings of the massXpert program. This manual is not a ``crash course'' in biochemistry.
2.1 Polymers? Where? Everywhere! #
Indeed, polymers are everywhere. If you ask somebody to show you something polymeric, he/she will point you at the first plastic object in the vicinity. Right, plastic materials are made of hydrocarbon polymers. We also have many different polymers in our body. Proteins are polymers, complex sugars are polymers, DNA (the so-called “molecule of heredity” is a huge polymer. There are polymers in wine, in wood... Where? Everywhere!
The Oxford Advanced Learner's Dictionary of Current English gives for polymer the following definition: —“natural or artificial compound made up of large molecules which are themselves made from combinations of small simple molecules”.
A polymer is indeed made by covalently linking small simple molecules together. These small simple molecules are called monomers, and it is immediate that a polymer is made of a number of monomers. A general term to describe the process that leads to the formation of a polymer is polymerization. It should be noted that there are many ways to polymerize monomers together. For example, a polymer might be either linear or branched. A polymer is linear if the monomers that are polymerized can be joined at most two times. The first junction links the monomer to an elongating polymer (thus making it the new end of the elongating polymer which, by the way, is longer than before by one unit) and the second junction links the new elongating polymer's end to another monomer. This process goes on until the reaction is stopped, the point at which the polymer reaches its finished state. A branched polymer is a polymer in which at least one monomer is able to contract more than two bonds. It is thus clear that a single monomer linked three times to other monomers will yield a “T-structure”, which is nothing but a branched structure.
In the following sections we'll describe a number of different kinds of polymers. Each time, they will be described by initially detailing the structure of their constitutive monomers; next the formation of the polymer is described. At each step we shall try to set forth each polymer characteristics in such a manner as to introduce the way massXpert “thinks polymers” and to introduce specialized terminologies. Once the basic chemistries (of the different polymers) have all been described, we will enter a more complex subject that is of enormous importance to the mass spectrometry specialist: polymer chain disrupting chemistry. We shall see that this terminology actually involves two kinds of chemistries: cleavage, on the one hand, and fragmentation, on the other hand.
While massXpert is basically oriented to linear single-stranded polymer chemistries, it can also be used to simulate highly complex polymer chemistries. Biological polymers are the main focus of this manual, however all the concepts described here may be applied with no modification to synthetic polymer chemistries.
2.2 Various Biopolymer Structures #
Biopolymers are amongst the most sophisticated and complex polymers on earth and it certainly is not a mistake to take them as examples of how monomers (be these complex or not) can assemble covalently into life-enabling polymers. In this section we will visit three different polymers encountered in the living world: proteins, nucleic acids and polysaccharides. We shall be concerned with 1) the monomers' structure, 2) the polymerization reaction and 3) the final end-capping reaction responsible for putting the polymer in its finished state.
2.2.1 Proteins #
These biopolymers are made of amino acids. There are twenty major amino acids in nature, and each protein is made of a number of these amino acids. The combinations are infinite, providing enormous diversity of proteins to the living world.
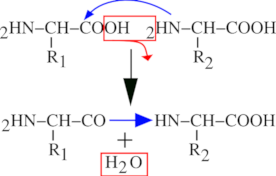
A protein is a polar polymer: it has a left end and a right end, and polymerization actually occurs from left to right (from N-terminus to C-terminus, see below). Figure 2.1, “Peptidic bond formation by condensation.” shows that the chemical reaction at the basis of protein synthesis is a condensation. A protein is the result of the condensation of amino acids with each other in an orderly polar fashion. A protein has a left end, called N-terminus; amino terminal end and a right end, called C-terminus; carboxyl terminal end. The left end is an amino group (2HN–) corresponding to the non-reacted amino group of the amino acid. Upon condensation of a new amino acid onto the first one, the carboxyl group of the first amino acid reacts with the amino group of the second amino acid. A water molecule is released, and the formation of an amide bond between the two amino acids yields a dipeptide. The right end of the dipeptide is a carboxyl group (–COOH) corresponding to the un-reacted carboxyl group of the last amino acid to have “polymerized in”.
The bond formed by condensation of two amino acids is an amide bond, also called—in protein chemistry—a peptidic bond. The elongation of the protein is a simple repetition of the condensation reaction shown in Figure 2.1, “Peptidic bond formation by condensation.”, granted that the elongation always proceeds in the described direction (a new monomer arrives to the right end of the elongating polymer, and elongation is done from left to right).
Figure 2.1: Peptidic bond formation by condensation. #
Now we should point at a protein chemistry-specific terminology issue: we have seen that a protein is a polymer made of a number of monomers, called amino acids. In protein chemistry, there is a subtlety: once a monomer is polymerized into a protein it is no more called a monomer, it is called a residue. We may say that a residue is an amino acid less a water molecule.
From what we have seen until now, we may define a protein this way: —“A protein is a chain of residues linked together in an orderly polar fashion, with the residues being numbered starting from 1 and ending at n, from the first residue on the left end to the last one on the right end”. This definition is still partly inexact, however. Indeed, from what is shown in Figure 2.2, “End capping chemistry of the protein polymer.”, there is still a problem with the extremities of the residual chain: what about the amino group on the left end of a protein (the amino group sits right onto the first amino acid of the protein), and what about the carboxyl group of the right end of a protein (the carboxyl group sits right onto the last amino acid of the protein)? Because these groups lie at the extremities of the residual chain, they remained unreacted during the polymerization process. But because we are simulating a residual chain using residues and not amino-acids, we still need to put the residual chain in its finished state: by capping the left end with a proton cap (so as to complete the amino group) and the right end with a hydroxyl cap (so as to complete the carboxyl group). The capping of the residual chain extremities ensures that the polymer is in its finished state, and that it cannot be elongated anymore. The proton is the left cap of the protein polymer and the hydroxyl is the right cap of the protein polymer.
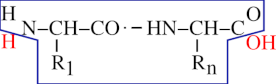
A protein is made of a chain of residues and of two caps. The left cap is the N-terminal proton and the right cap is the C-terminal hydroxyl. Altogether, the residual chain (enclosed here in the blue polygon) and both the H and OH red-colored caps do form a complete protein polymer in its finished state.
Figure 2.2: End capping chemistry of the protein polymer. #
Now comes the question of unambiguously defining the structure of a protein. It is commonly accepted that the simple ordered sequence of each residue code in the protein, from left to right, constitutes an unambiguous description of the protein's primary structure (that is its sequence). Of course, proteins have three-dimensional structures, but this is of no interest to a program like massXpert, which is aimed at calculating masses of polymers. To enunciate unambiguously the sequence of a protein, one would use a symbology like this:
Using the 3-letter code of the amino acids:
Ala Gly Trp Tyr Glu Gly Lys
Using the 1-letter code of the amino acids:
A G W Y E G K
Alanine is thus the residue 1 and Lysine is the last residue (n = 7)
2.2.2 Nucleic Acids #
These biopolymers are more complex than proteins, mainly because they are composed of monomers (nucleotides) that have three different chemical parts, and because those parts differ in DNA and RNA. A nucleotide is the nucleic acid's brick: a nucleotide consists of a nitrogenous base combined with a ribose/deoxyribose sugar and with a phosphate group. There are two different kinds of nucleic acids: deoxyribonucleic acid (DNA, the sugar is a deoxyribose) and ribonucleic acid (RNA, the sugar is a ribose). DNA is most often found in its double stranded form, while RNA is most often found in single strand form. There are four nitrogenous bases for each: Adenine, Thymine, Guanine, Cytosine for DNA; in RNA only one of these bases changes: Thymine is replaced by Uracile. As for proteins, nucleic acids are polar polymers: the polymerization process is polar, from left to right (sometimes left is up and right is down in certain vertical representations found mainly in textbooks).
This manual is not to teach biochemistry, which is why the structure of the monomers is not described in atomic detail. However, since it is important to understand how the polymerization occurs, Figure 2.3, “Phosphodiester bond formation by esterification.” represents the polymerization reaction mechanism between a nucleotide and another one, to yield a dinucleotide. That reaction is a trans-esterification. A nucleic acid has a left end—5' end; often this end is phosphorylated—and a right end—3' end; hydroxyl end. The trans-esterification reaction is the attack of the phosphorus of the new (deoxy)nucleotide triphosphate by the 3'OH of the right end of the elongating nucleotidic chain. Upon trans-esterification, an inorganic pyrophosphate (PPi) is released, and the formation of a phosphodiester bond between the two nucleotides yields a dinucleotide. The elongation of the nucleic acid polymer is a simple repetition of this esterification reaction so that the chain growth is always in the 5'→3' direction. This is achieved in the living cells by what is called the 5'→3' polymerase enzymatic activity.
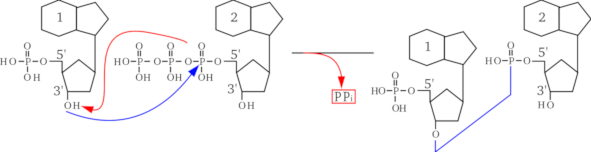
The arriving monomer (on the right) has its triphosphate on the 5' carbon of the sugar esterified by nucleophilic attack of the first phosphorus by the alcohol function beared by the 3' carbon of the (deoxy)ribose sugar ring of the left monomer. The bond that is formed is a phosphodiester bond, with release of a pyrophosphate group (PPi). Note that the sugar and nitrogenous bases are schematically represented in this figure.
Figure 2.3: Phosphodiester bond formation by esterification. #
The conventional representation of a nucleic acid involves showing the 5' end on the left, and the 3' end on the right, horizontally. Sometimes, to clearly indicate that the left end is phosphorylated, while the right end is not, the ends are indicated as ``5'P'' and ``3'OH''. Figure 2.4, “End capping chemistry of the nucleic acid polymer.” shows a simple way to formalize what a nucleic acid polymer is. The molecule represented on the left is the “monomer” in the sense that the polymer is made of n monomers. On the right side of that figure, the polymer made of n monomers is shown as a residual chain (inside the blue polygon box) that got capped with OH on its left end and H on its right end (red-colored atoms). Thus, in the case of the nucleic acid polymers, the left cap is a hydroxyl and the right cap is a proton. This anecdotically happens to be the exact converse of what was described earlier for proteins.
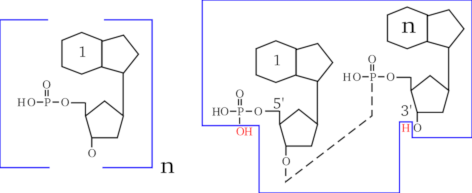
A nucleic acid is made of a chain of nucleotides (left formula) and of two caps. The left cap is the hydroxyl group that belongs to the terminal phosphate of the 5' carbon of the sugar. The right cap is the proton that belongs to the hydroxyl group of the 3' carbon of the sugar ring (right formula). Altogether, a finished nucleic acid polymer is made of the nucleotidic chain (enclosed here in the blue polygon), made of the repetitive elements (one of which is shown on the left), and of the two caps (red-colored OH and H, out of the box on the right).
Figure 2.4: End capping chemistry of the nucleic acid polymer. #
Now comes the question of unambiguously defining the structure of a nucleic acid. It is commonly accepted that the listing of the named nitrogenous bases in the nucleic acid—from left (5' end) to right (3' end)—constitutes an unambiguous description of the nucleic acid sequence. To enunciate the sequence of a gene, one would use a symbology like this:
for a DNA, using the 1-letter code of the nitrogenous bases:
A T G C A G T C
for an RNA, using the 1-letter code of the nitrogenous bases:
A U G C A G U C
Adenine is thus the base 1 and Cytosine is the last base (n = 8)
2.2.3 Saccharides #
These biopolymers are certainly amongst the most complex ones in the living world. This is mainly due to the fact that saccharides are usually heavily modified in living cells with a huge variety of chemical modifications. Furthermore, the ramifications in the polymer structure are more often the normal situation than not. Interestingly, these molecules are first thought of as the “fuel” for the cell, which is certainly far from being total nonsense, but it is also undoubtful that their structural role is extremely important (often in combination with proteinaceous material). Another interesting aspect of their ability to form complex structures is their use as “key” systems for identification processes: a number of complex sugars are located on the cell walls and provide “recognition patterns” for the other cells to deal with…
Nonetheless, the general picture is not that complex, if the way monomers are polymerized together is the only concern (which is the case in this manual). As far as we are concerned, in fact, the polymerization mechanism is a simple condensation (much like what has been described for proteins), yielding a sugar bond. Indeed, some people use the same terminology: a monomeric sugar becomes a residue once polymerized in the saccharidic chain. There are two main different kinds of sugars: pentoses (in C5) and hexoses (in C6); it should be noted, however, that there is a variety of other common molecules, like sialic acids, heptoses…
Like already seen for proteins and nucleic acids, a saccharidic polymer is polar: it has a left end and a right end. The terminology regarding the ends of a saccharidic polymer is rather unexpected at first sight: the left end is said to be the non-reducing end while the right end is said to be the reducing end. Historically this was observed with monosaccharides (also called monoses), which reduced cupric (Cu2+) ions, thus getting oxydized themselves on the carbonyl (when in the open ring aldehydic form).
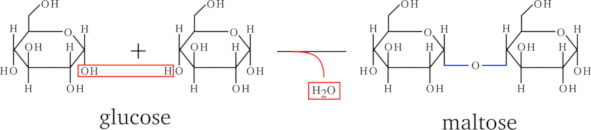
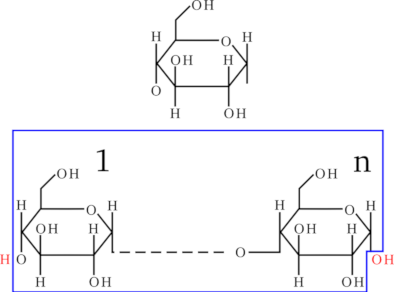
Figure 2.5, “Osidic bond formation by condensation.” shows the polymerization reaction between a sugar and another one (2 glucose monomers, actually), to yield a maltose disaccharide. The polymerization mechanism is a simple condensation. The elongation of the saccharidic polymer is a simple repetition of this condensation reaction so that the chain growth is always in the same orientation, from the non-reducing end to the reducing end. The conventional representation of a polysaccharide involves showing the non-reducing end on the left, and the reducing end on the right, horizontally. Figure 2.6, “End capping chemistry of the polysaccharidic polymer.” shows a simple way to formalize what a saccharidic polymer is. The top formula is the representation of the monomer. The bottom formula represents a polysaccharide, with the repetitive elements boxed (there are n monomers polymerized). The atoms shown in red (outside the boxed repetitive elements) are the saccharidic polymer caps. Thus, we see clearly that in the case of polysaccharides, the left cap is a proton and the right cap is a hydroxyl. This anecdotically happens to be identical to proteins and the exact converse of what we described previously for nucleic acids.
Figure 2.5: Osidic bond formation by condensation. #
Figure 2.6: End capping chemistry of the polysaccharidic polymer. #
Now comes the question of unambiguously defining the structure of a saccharidic polymer. It is commonly accepted that the simple ordered sequence of the named monoses in the saccharidic polymer, from left (non-reducing end) to right (reducing end), constitutes an unambiguous description of the glycan sequence. To enunciate the sequence of a glycan, one would use a symbology like this:
Using a 3-letter code:
Ara Gal Xyl Glc Hep Man Fru
Arabinose is thus the monose 1 and Fructose is the last monose (n = 7)
Incidentally, this is where the ability of massXpert to handle monomer codes of non-limited length comes in handy!
2.3 To Sum Up #
We made a rapid overview of the three major polymers in the living world. A great many other polymers exist around us. Table 2.1, “Quick comparison of three biopolymers with examples of monomers” tries to sum up all the informations gathered so far. Note that the formulæ given for the monomers are the “residual” ones. For example, the formula of the glycyl residue corresponds to the formula of the Glycine monomer less one molecule of water. Many synthetic polymers are much simpler than the ones we have rapidly reviewed, and it should be clear that, if massXpert can deal with the complex biopolymers described so far, it certainly will be very proficient with less complex synthetic polymers. Describing the formation of polymers is one thing, but we also have to describe how to disrupt polymers. This is what we shall do in the next section.
Table 2.1: Quick comparison of three biopolymers with examples of monomers #
| polymer | name | code | formula | left cap | right cap |
| protein | H | OH | |||
| Glycine | G | C2H3O1N1 | |||
| Alanine | A | C3H5O1N1 | |||
| Tyrosine | T | C9H9O2N1 | |||
| nucleic acid | OH | H | |||
| Adenine | A | C10H12O5N5P1 | |||
| Cytosine | C | C9H12O6N3P1 | |||
| saccharide | OH | H | |||
| Arabinose | Ara | C5H8O4 | |||
| Heptose | Hep | C7H12O8 |
2.4 Polymer Chain Disrupting Chemistry #
The “polymer chain disrupting chemistry” was mentioned earlier as a complex subject that was of enormous importance to the mass spectrometrist. This is why that subject will be treated in a pretty thorough manner. First of all it should be noted that a chemical modification of a polymer does not necessarily involve the perturbation of the chain structure of the polymer. Here, however, we are concerned specifically with a number of chemical modifications that yield a polymer chain perturbation; cleavage and fragmentation:
Cleavages. These are chemical processes by which a cleaving agent will act directly on the polymer chain making it fall into at least two separated pieces (the oligomers). As a result of the cleavage reaction, groups originating in the cleaving molecule remain attached to the polymer at the precise cleavage location;
Fragmentations. These are chemical processes by which the polymer structure is disrupted into separated pieces (the fragments) mainly because of energy-dependent electron doublet rearrangements leading to bond breakage.
2.4.1 Polymer Cleavage #
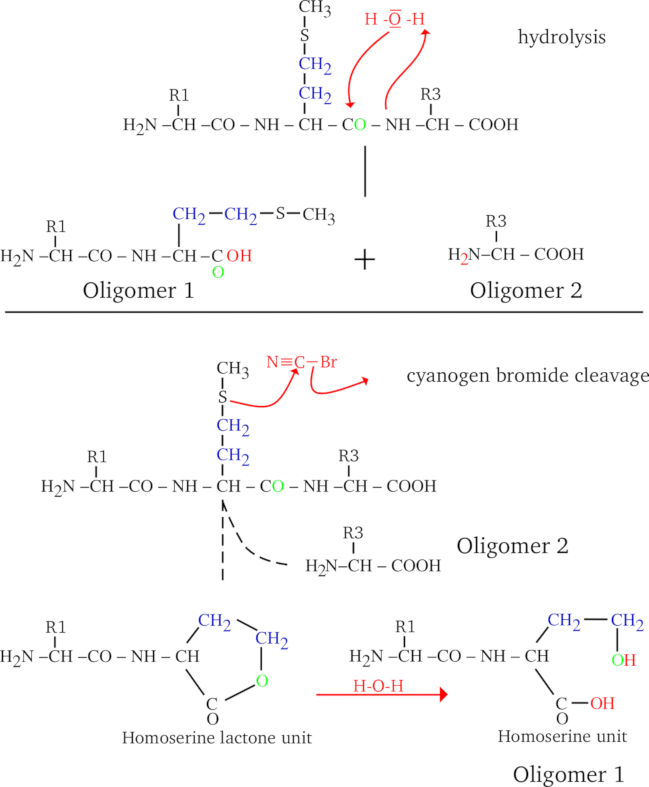
We said above that, upon cleavage of a polymer, the cleaving molecule reacts with it, and by doing so directly or indirectly “dissolves” an inter-monomer bond. A polymer cleavage always occurs in such a way as to generate a set of true polymers (smaller in size than the parent polymer, evidently, which is why they are called oligomers). Indeed, let us take the example shown in Figure 2.7, “Protein cleavage by water and cyanogen bromide”, where a tripeptide (a very little protein, containing a methionyl residue at position 2) is submitted either to a water-mediated cleavage (hydrolysis, upper panel) or to a cyanogen bromide-mediated cleavage (lower panel). The two cases presented in this figure are similar in some respects and different in others:
In the first case the molecule that is responsible for the cleavage is water, while in the second case it is cyanogen bromide;
In both cases the bond that is cleaved is the inter-monomer bond (in protein chemistry this is a peptidic bond);
In both cases the Oligomer 2 has the same structure;
The structures of the Oligomer 1 species differ when produced using water or cyanogen bromide as the cleaving molecule.
The difference between hydrolysis and cyanogen bromide cleavage is in the generation of the Oligomer 1 species: the cyanogen bromide cleavage has a side effect of generating a homoserine as the right end monomer of Oligomer 1, while hydrolysis generates a genuine methionine monomer. This is because water reverses in a very symmetrical manner what polymerization did (hydrolysis is the converse of condensation), while cyanogen bromide did some chemical modification onto the generated Oligomer 1 species.
Figure 2.7: Protein cleavage by water and cyanogen bromide #
Nonetheless, the reader might have noted that—interestingly—all the four oligomers do effectively have their left cap (a proton) and their right cap (the hydroxyl). This means that in both water- and cyanogen bromide-mediated cleavages, all the generated oligomers are indeed true polymers in the sense that: 1) they are a chain of monomers (modified or not) and 2) they are correctly capped (i.e. they are polymers in their finished state). This is important because it is the basis on which we shall make the difference between a cleavage process and a fragmentation process. Thus, the massXpert definition of an oligomer might be: an oligomer is a polymer (of at least one monomer) in its finished state that was generated upon cleavage of a longer polymer.
When the polymer cleavage reaction precisely reverses the reaction that was performed for the same polymer's synthesis, there is no special difficulty. But when the cleavage reaction modifies the substrate, then this should be carefully modelled. How? To answer this question we might start by comparing the two different Oligomer 1 species that were yielded upon the water-mediated and the cyanogen bromide-mediated cleavage reactions: “the hydrolysis-generated Oligomer 1 is equal to the cyanogen bromide-generated Oligomer 1 +S1 +C1 +H2 -O1”; this is a big difference! The observations we did so far might be worded this way: Whenever a protein undergoes a cyanogen bromide-mediated cleavage, the “-C1H2S1+O1” chemical reaction should be applied to the resulting oligomers if and only if they have a methionine monomer at their right end. In massXpert's jargon, this logical condition is called a cleavage rule (described later; see Section 3.2.4, “ The cleavage specifications”).
Well, all this sounds reasonable. But what about the “normal” case, when the cleavage is done using water? Nothing special: the mass of the oligomer is calculated by summing the mass of each monomer in the oligomer (since the monomers are not modified, this is easily done) and the masses corresponding to the left and right caps (these are defined in the polymer chemistry definition; in our present case it would be a proton on the left end, and a hydroxyl on the right end). In this way, the oligomer complies with its definition, which states that it is a faithful polymer made of monomers and that it is in its finished state.
Yes, but then how will massXpert manage to calculate the mass of the modified oligomer, like our Oligomer 1 in the case of the cyanogen bromide-mediated cleavage? Simple enough: in a first step it does exactly the same way as for the unmodified oligomer. Next, each oligomer is checked for presence or absence of a methionine residue on its right end. If a methionine is found, the mass corresponding to the “-C1H2S1+O1” chemical reaction is applied. And that's it.
In the previous cyanogen bromide example, the logical condition was involving the identity of the oligomers' right end monomer, but other examples can involve not the right end monomer, but the left end monomer, if some chemical modification was to occur to the monomer sitting right of the cleavage location. In this case the user would have to analyse the situation and provide massXpert with the proper chemical reaction by stating something analog to: if and only if they have a Xyz monomer at their left end. This introduction to polymer cleavage abstraction should be enough to later delve into the cleavage specification definition as massXpert conceives it and that is thoroughly detailed at Section 3.2.4, “ The cleavage specifications”.
2.4.2 Polymer Fragmentation #
In a fragmentation process, the bond that is broken is not necessarily the inter-monomer bond. Indeed, fragmentations are oft-times high energy chemical processes that can affect bonds that belong to the monomers' internal structure. This is one of the reasons why fragmentations do differ from cleavages: they are specific of the polymer type in which they occur. Hydrolyzing a protein and an oligosaccharide is just the same process, from a chemical point of view. But fragmenting a protein or an oligosaccharide are truly different processes because the way that the fragmentation happens in the polymer sequence is so much dependent on the nature of each monomer that makes it.
Another peculiarity of the fragmentations, compared with the cleavages that were described above, is the fact that there is no cleaving molecule starting the process. Instead, a fragmentation process is often initiated by an intra molecular electron doublet rearragement that propagates more or less in the polymer structure to eventually break it. Fragmentations are mainly a gas phase process, not some reaction that happens in solution as a result of putting in contact the polymer and some reagent. It is precisely because no cleaving molecule is involved in the fragmentation process that the fragments are not necessarily capped like a normal polymer should be; and this is another really important difference between cleavage and fragmentation. The following examples should illustrate these concepts: protein and nucleic acid fragmentation.
2.4.2.1 Protein Fragmentation #
There is a pretty important number of different kinds of fragments that can be generated upon fragmentation of peptides. We are going to detail the most common ones; the user is invited to use the massXpert' fragmentation-specification grammar to add less frequent (or newly discovered) fragmentation types. Note that the fragmentation schemes below apply to positively-charged precursor ions. To compute the product ions' masses obtained in negative mode fragmentation experiments, then, simply remove as many protons as required. For example, to switch from a fragment positively charged once (+H), then remove a first proton to go back to the uncharged state and then remove another proton to yield the deprotonated (thus singly negatively charged) ion product. The requirement to be able to computed masses for the positively- and negatively-charged ion products imposes a specific way to defined fragmentation specifications in the XpertDef module (to be detailed later in this manual).
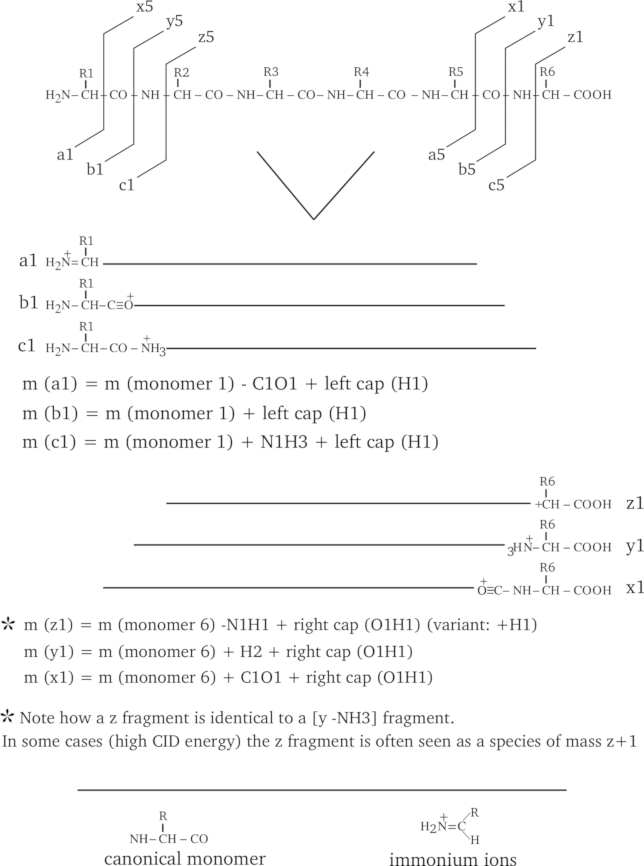
An hexapeptide is fragmented in the seven most widely encountered manners, such as to generate a, b, c, x, y, z and immonium fragment ions. The figure illustrates the position of the cleavage for each kind of fragment (exemplified using the case of the smallest fragment possible) and the mass calculation method is described for each fragment kind; consider that each fragment bears only one positive charge.
Figure 2.8: Protein fragmentation patterns most widely encountered. #
As can be seen from Figure 2.8, “Protein fragmentation patterns most widely encountered.”, the fragmentations do generate fragments of three categories: the ones that include the left end of the precursor polymer (a, b, c), the ones that include the right end of the precursor polymer (x, y, z), and finally the special case in which the fragment is an internal fragment, like the immonium ions. When looking at the fragmentations described in the figure it becomes immediately clear why a fragmentation cannot be mistaken for a cleavage: the ionization of the fragment is not necessarily due to the captation of a proton by the fragment. Furthermore, we can also see that a fragmentation is not a cleavage because the fragment that is generated is absolutely not necessarily what we call a polymer, in the sense that the fragment might not be capped the same way as the precursor polymer is (that is, the fragment is not in its finished polymerizaton state).
The two observations above should make clear to the reader that calculating masses for fragments is a more difficult process than what was described above for the oligomers. Indeed, while it was simple to calculate the mass of an oligomer (by simply adding the masses of its constitutive monomer units, plus the left and right caps, plus ionization), here there is no chemical formalism generally applicable to all the fragment types. This is why the specification of the fragmentation is left to the user's responsibility.
By looking at Figure 2.8, “Protein fragmentation patterns most widely encountered.”, the reader should have noticed that the fragment naming scheme takes into consideration the fact that the fragment bears the left or the right end of the precursor polymer (or none, also). Indeed, the numbering of fragments holding the left end of the precursor polymer sequence begins at the left end, and for fragments that hold the right end, at the right end. Thus the third fragment of series a (a3) would involve monomers [1→] and the third fragment of series y (y3) would involve monomers [6→] (in the figure, these left-to-right and right-to-left directions are symbolized using arrows). Therefore, it should appear to the reader how important—when specifying a fragmentation—it is to clearly indicate from which end of the precursor polymer the fragment is generated (in massXpert's jargon this is “LE” for left end, “RE” for right end and “NE” for no end). massXpert knows what action it should take when it encounters one of these three specifications; for example, if a “LE” specification is found for a given fragmentation specification, massXpert adds to the fragment's mass the mass corresponding to the left cap of the precursor polymer.
The mass calculations for the different fragment product ion types is shown in Figure 2.9, “Peptide product ion mass calculation for the different ion series”.
Figure 2.9: Peptide product ion mass calculation for the different ion series #
If we take the a fragment series, the Figure 2.8, “Protein fragmentation patterns most widely encountered.” indicates that the fragments include the left end and that their last monomer lacks its carbonyl group (see, on top of Figure 2.8, “Protein fragmentation patterns most widely encountered.”, that the a1 arrow goes between the CαH and the CO of monomer 1). So we would say that each fragment of the a series should be challenged with the following chemical treatments: 1) addition of the mass corresponding to the left cap (proton), 2) removal of the mass corresponding to the lacking CO group. This way we have the mass of fragment a1. If we were interested in the fragment a4 we would have summed the masses of monomers 1 to 4, added the mass of the left cap, and finally removed the mass of a CO.
For the x series of fragments we do not add the left cap anymore, but replace it with the right cap, since the fragments hold the right end of the precursor polymer. Note also that the numbering of the monomers using the variable i in the following mathematical expressions goes from right to left (contrary to what happened for the a, b, c fragment series. All the fragments that hold the precursor polymer right end are numbered this way, so this applies to fragments x, y, z.
In low energy CID, the z fragments are expressed in a distinct way, when compare to high energy CID.
In the immonium fragment series, the fragments are internal fragments in the sense that they do not hold neither of the two precursor polymer's ends. massXpert understands that the user is speaking of this kind of fragment when the “from which end” piece of data —in the fragmentation specification—states “NE” instead of “LE” or “RE” (see Section 2.4.2, “Polymer Fragmentation”). The mass calculation for these fragments does not take into account the monomers surrounding the one for which the calculation is done. The mass for an immonium ion —at position i in the precursor polymer—will be the mass of the monomer at position i, less the mass of a CO, plus the mass of a proton.
2.4.2.2 Nucleic Acids Fragmentation #
The fragmentations that can be obtained with nucleic acids are numerous and it is more complicated than with proteins to describe them fully. The main reason for this is that there are a big number of fragmentation combinations because of the loss of nitrogenous bases from the skeleton. The mechanisms by which this loss happens are fairly complex, and I am not going to detail any of them. Figure 2.10, “DNA fragmentation patterns most widely encountered.” shows the most common fragmentations (without taking into consideration the potential loss of bases). An example of fragment is given for each fragment series (pretty the same way as we did before for proteins). Note that the fragment representations are aimed at helping the reader to figure out what the product ion is, not taking into account where the negative charge lies on the fragment, since this charge can float around at every de-protonatable group. All the fragments shown bear one and one only charge.
Another remark pertaining to the ionization mode of the ion products: the fragmentation schemes below apply to negatively-charged precursor ions (by loss of a proton, typically). To compute the product ions' masses obtained in positive mode fragmentation experiments, then, simply add as many protons (or any other cationic ionization agent) as required. For example, to switch from a fragment negatively charged once (-H), then add a first proton to go back to the uncharged state and then add another proton to yield the monoprotonated (thus singly positively charged) ion product. The requirement to be able to computed masses for the positively- and negatively-charged ion products imposes a specific way to defined fragmentation specifications in the XpertDef module (to be detailed later in this manual).
The reader might have noticed at the bottom of Figure 2.10, “DNA fragmentation patterns most widely encountered.” that a provision is made in the case the fragmented molecular species are not 5' end-phosphorylated but 5' end-hydroxylated. Indeed, the canonical monomer is such that, upon polymerization and left capping, the 5' end is phosphorylated. However, oft-times the oligonucleotides are synthesized chemically without the 5' end phosphate group, thus ending in hydroxyl. This special case should be accounted for by applying to all the fragments that bear the left end of the precursor polymer the following chemical reaction: -HPO3. This chemical reaction should be applied in addition to the chemical reaction that yields the fragment per se.
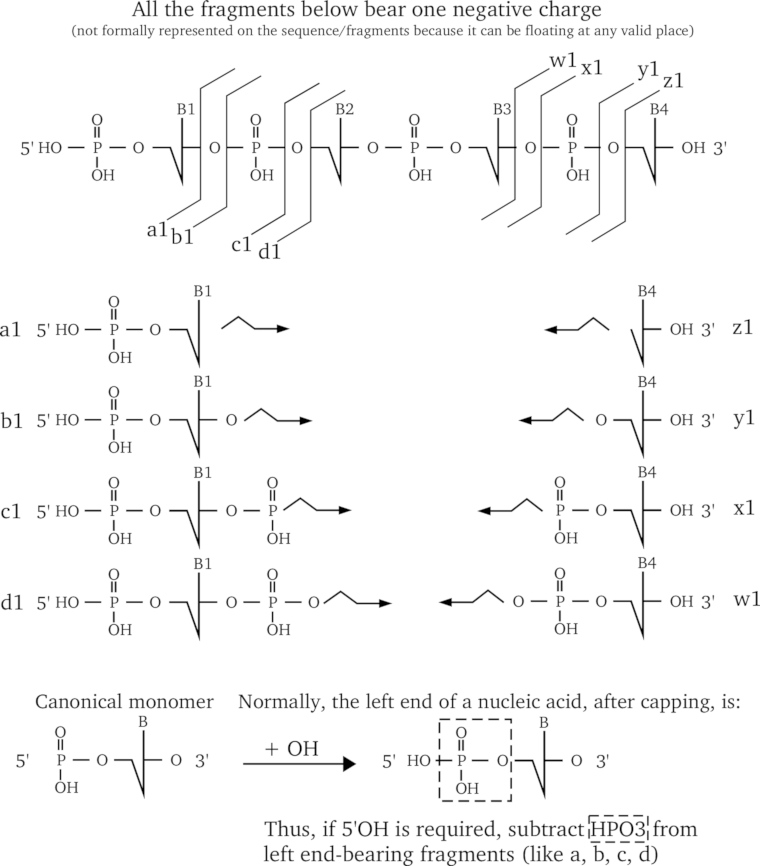
A short DNA sequence is fragmented in the eight most widely encountered manners, such as to generate a, b, c, d, w, x, y, z fragment ions. The figure illustrates the position of the cleavage for each kind of fragment (exemplified using the case of the smallest fragment possible). and the mass calculation method is described for each fragment kind; considering that each fragment is protonated only once (+1).
Figure 2.10: DNA fragmentation patterns most widely encountered. #
Exactly as done earlier for the protein fragments, the mathematical expressions used to calculate the mass of different series of nucleic acid fragments are provided in Figure 2.11, “Nucleic acids product ion mass calculation for the different ion series”. In these calculations it is assumed that the left end of the precursor polymer is phosphorylated (5'P) and the reader should bear in mind that this precise phosphate might itself be expelled by the fragmentation. The fragment naming schemed detailed earlier for proteins applies to nucleic acids in the very same manner.
Figure 2.11: Nucleic acids product ion mass calculation for the different ion series #
In the a fragment series, the fragments most often appear with base loss. There are also a variety of fragments for which a base is lost.
2.4.2.3 More Complex Patterns Of Fragmentation #
Before finishing with fragmentations, it is necessary to describe a powerful feature of the fragmentation specification grammar available in massXpert. This feature was required for the fragmentation of oligosaccharides and also sometimes for proteins. When the fragmentation (the bond breakage reaction itself) occurs at the level of certain monomers, it might be necessary to be able to specify some particular chemistry that would arise on the monomer in question.
We have seen in the cleavage documentation that, upon cleavage of a protein sequence with cyanogen bromide, for example, a particular chemical reaction had to be applied to the oligomers that were generated with a methionine monomer as their right end monomer. Well, in a fragmentation specification it is possible to apply comparable chemical reactions but in a more thorough manner. Indeed, while in the cleavage it was possible to say something like: —“Apply a given chemical reaction to the oligomer if the right end monomer is Xyz”, in the fragmentation the logical condition can be bound not only to the identity of the currently fragmented monomer, but also (optionally) to the identity of the previous and/or next monomer in the precursor polymer sequence. For example: —“Apply a given chemical reaction if fragmentation occurs at the level of ‘Xyz’ monomer only if it is preceded by a ‘Yxz’ monomer and followed by a ‘Zyx’ monomer”.
These logical conditions are called fragmentation rules. A fragmentation specification can hold as many rules as necessary. All of this is described in great detail at Section 2.4.2, “Polymer Fragmentation”.
2.4.2.4 To Sum Up #
To sum up all what we have seen so far with polymer chain disrupting chemistries:
Fragments are never ionized automatically; ionization (gain/loss of a charged group) is necessarily integrated in the fragmentation specification.
A polymer sequence gets fragmented into fragments when a bond breakage occurs, without the help of any exterior molecule, at any level of the polymer structure, with no limitation to the inter-monomer bond; monomer-specific chemical reactions can be modelled into the fragmentation specification using any number of fragrules;
Oligomers are automatically capped—on both ends—using the rules described in the precursor polymer's definition;
Fragments are capped automatically only—on the end they hold, if any—using the rules described in the precursor polymer's definition;
Oligomers are automatically ionized (if required by the user) using the rules described in the precursor polymer's definition;
Fragments are never ionized automatically; ionization (gain/loss of a charged group) is necessarily integrated in the fragmentation specification.