massXpert2 User Manual
6 XpertMiner: a Data Miner
XpertMiner is a module that has been conceived as a repository of functionalities aimed at analyzing mass data. The data to be subjected to mining can originate from:
massXpert-based simulations, like polymer cleavage, oligomer fragmentation or arbitrary mass searches;
An export of a mass list from the mass spectrometer software;
Any mass data that might have been processed outside of massXpert and that need to be reimported in XpertMiner.
6.1 XpertMiner Invocation #
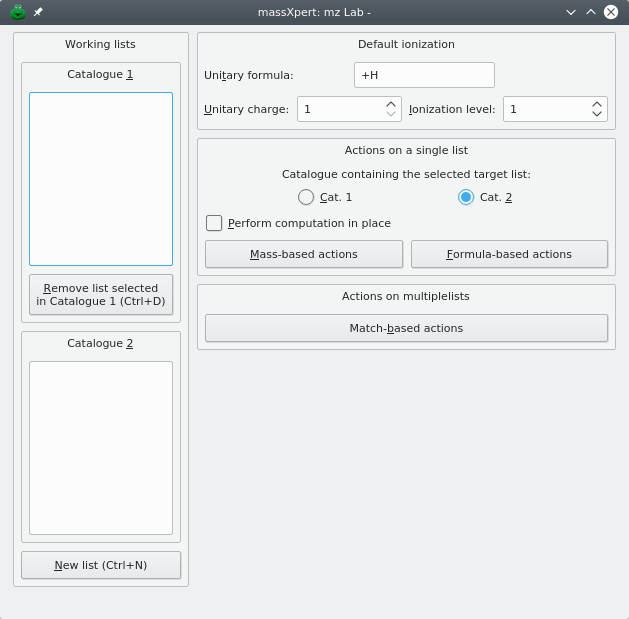
The XpertMiner module is easily called by pulling down the menu item from the massXpert program's menu. Clicking on → will open the mzLab window, as represented in Figure 6.1, “mzLab window”.
6.2 mzLab: Mining m/z Ratios #
The features available in this laboratory operate on lists of m/z values in the form of (m/z,z) pairs. The mass of the ion is represented by “m”, while “z” is the charge of the ion. With the two data in the pair, the m/z ratio and the z charge, and knowing the ionization rule that ionized the analyte in the first place, it is possible to perform any mass calculation on the (m/z,z) pair.
The mzLab window is represented in Figure 6.1, “mzLab window”. This window is divided into a number of distinct parts:
The left part (Working lists) contains two list widgets which will hold the names of the different working m/z lists. We call these lists “catalogues” in the following text;
The Default ionization groupbox widget contains the ionization rule that is to be assumed when working on (m/z,z) pairs. If you are unsure about this concept, please read section Section 3.2, “ The polymer chemical entities”.
The Actions on a single list groupbox widget holds a number of mining actions to be performed on a single list that is identified by being selected either in the Cat. 1 or in the Cat. 2 catalogue of available m/z lists.
When performing computations that modify the m/z values in the list, if Perform computation in place is checked, then the new m/z values will replace the former ones. Otherwise, the program will ask for a new m/z list name as a new list is created to hold the new m/z values resulting from the computation.
There are two main kinds of computations that might be performed against a single m/z list:
Mass-based actions rely on masses or m/z values to perform computations;
Formula-based actions rely on formulæ to perform computations;
The Actions on multiple lists groupbox widget allows one to perform actions that use two lists, for example matching masses (or m/z ratios) in two lists with a given tolerance.
Figure 6.1: mzLab window #
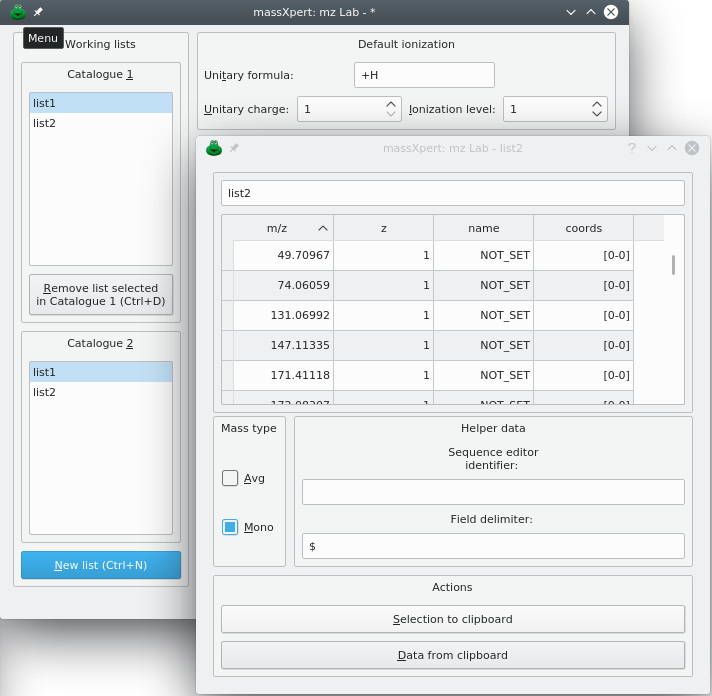
6.3 Creating a New Input m/z List #
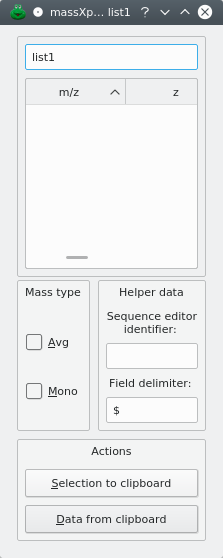
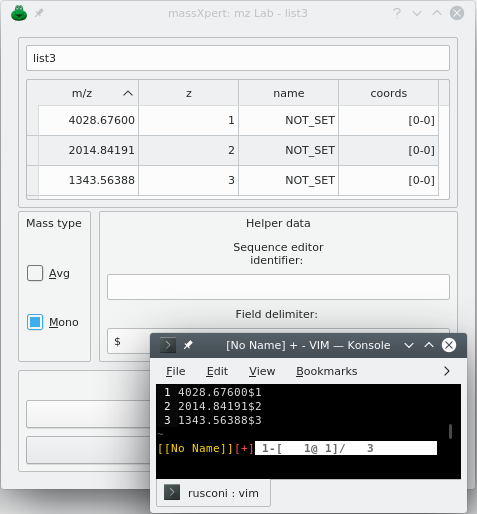
In order to be able to use the mzLab, it is necessary to create at least one list of (m/z,z) pairs, which is referred to by “input m/z list”, for short. To create a new input m/z list, you click New list. An input dialog window let's you enter the name of the new list. The new input m/z list dialog window shows up empty like in Figure 6.2, “m/z list's empty input m/z list dialog window.”. That kind of list is actually a table view widget that is embedded in a dialog window. The first column of the table view widget holds the m/z value, and the second column, the z value. Optionally, the name of the corresponding oligomer and its coordinates in the polymer sequence can be shown, depending on how data have been imported into the m/z list (see below).
Figure 6.2: m/z list's empty input m/z list dialog window. #
Tip
The list name entered by the user at creation time will be used to refer to that list in the two catalogues.
6.3.1 Filling m/z Lists with Data #
Once a new input m/z list has been named and created, it is necessary to fill it with (m/z,z) pairs. This is performed via drag-and-drop or clipboard operations. There might be a number of different data sources to be used for filling the input m/z list, all reviewed in the following sections.
6.3.1.1 Importing Data from massXpert Results Windows #
Data from the various simulations available in massXpert include cleavage results, fragmentation results and mass search results, which all produce oligomers that are displayed in treeview widgets, as shown in Figure 4.13, “Polymer sequence cleavage window” or Figure 4.14, “Oligomer fragmentation window” or Figure 4.17, “Searching masses in a polymer sequence”.
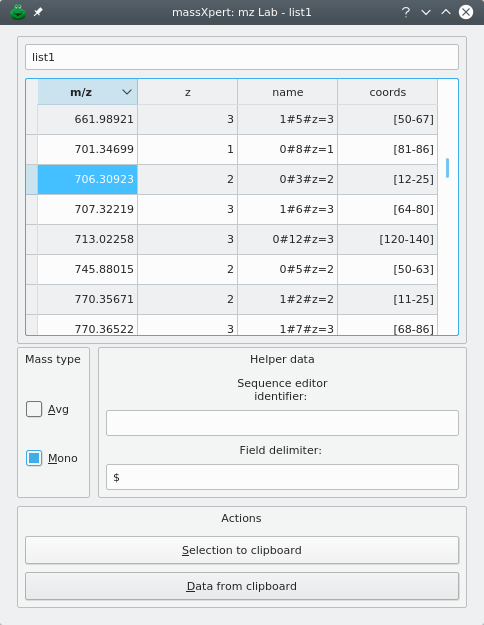
From these results windows, either select the oligomers of interest and export these data to the clipboard or perform a drag-and-drop operation to the m/z list area. Both ways produce identical results, as described in Figure 6.3, “m/z list's data-filled input m/z list dialog window”. One can see that the mass of the oligomers is set in the list, along with the charge, the oligomer name and, finally, the coordinates of the oligomer in the corresponding polymer.
Figure 6.3: m/z list's data-filled input m/z list dialog window #
6.3.1.2 Textual Data from Non-massXpert Results Windows, Without Charge #
The mass data might be first copied to the clipboard from other software and then imported in the m/z list by clicking Data from clipboard (a drag-and-drop operation would also work).
When the charge z is not present in the imported m/z list, then it is deduced from the ionization rule currently defined the the mzLab window (see background of Figure 6.4, “m/z list's (m/z) textual data-filled input m/z list dialog window.”).
Figure 6.4: m/z list's (m/z) textual data-filled input m/z list dialog window. #
6.3.1.3 Textual Data From Non-massXpert Results Windows, With z Charge Specified #
When the mass data copied to the clipboard do include the m/z ratio along with the z charge, the delimiter character needs to be known. This character must be set as the Field delimiter in the m/z list prior to clicking Data from clipboard to actually import the data. This way, the program knows how to parse the (m/z,z) pairs. A drag-and-drop operation from a graphical text editor would have produced the same results. The obtained list is shown in Figure 6.5, “m/z list's (m/z,z) textual data-filled input m/z list dialog window.”.
Figure 6.5: m/z list's (m/z,z) textual data-filled input m/z list dialog window. #
6.3.1.4 General Rules on Textual Mass Data Format #
The most detailed format that is supported is the following:
m/z <delim> charge <delim> name <delim> coordinates <delim>
In this syntax, the <delim> (field delimiter) is the “$” character. Any character might be used (including spaces). The delimiter character (or string) that is set in the m/z list window must be the same as the one defined in the window from where the data originate (when using the option to export the selected oligomers data to the clipboard).
For example, data can be formatted like this:
3818.05262$1$0#2#z=1$[3-39] 3834.05262$1$0#2#z=1$[3-39]
The compulsory datum (that is, the imported datum, either dragged and dropped or pasted from the clipboard), is the m/z ratio. The charge, name, coordinates fields are optional. If the charge is present, it will be taken into account while preparing the data for further use by the m/z list. If the charge is absent, it is deduced from the ionization rule currently defined in the mzLab window ( Figure 6.1, “mzLab window”).
Warning
If there is no charge value, then the other name and coordinates fields cannot be filled (or an error will result). The presence of the name and coordinates fields is optional. Note, however that the coordinates field is fundamental to be able to highlight the corresponding region in the XpertEdit sequence editor upon double-clicking of any given item in the m/z list. For this to be possible, the data must have been originated by drag and drop from a massXpert simulation results window or the m/z list window must have been connected to a polymer sequence editor window (see below).
6.3.2 Imposing the Mass Type: Mono Or Avg #
When dropping data—either from massXpert-driven simulations (cleavage, fragmentation or mass search) or from textual data originating from outside massXpert—it is necessary to inform the input m/z list of what kind of mass it is dealt with. That is, when dropping a line like “1234.56 1”, the question is: —“The m/z 1234.56 value is a monoisotopic m/z or an average m/z?” The type of the masses dropped in an input m/z list is governed by the two radio buttons labelled Mono and Avg. The one of the two radiobuttons that is checked at the moment the drop or the clipboard-paste occurs determines the type of the masses that are dealt with. It will be possible to check the other radio button widget once a first data drop occurred, but then the user will be alerted about doing so, as this has huge implications for the calculations to be performed later.
6.4 Working on One Input m/z List #
Once an input m/z list has been filled with data, it becomes possible to perform calculations on these data. Because there might be any number of input m/z lists open at any given time, it is necessary to identify the input m/z list onto which to perform these calculations. The selection of the input m/z list(s) is performed in two steps: first, by indicating in which catalogue the list of interest is currently selected (select either Cat.1 or Cat.3). Make sure a list name is currently selected in the proper catalogue.
6.4.1 Available Calculations #
There are a number of operations that might be performed, all of which are selectable in the Actions on a single list groupbox widget. The simulations are organized into two groups:
Formula-based actions which involve processing the input m/z lists with formulæ (that is, chemical entities represented using formulæ):
Apply formula will modify the m/z list by applying to all of its members the mass corresponding to the formula entered by the user. This is where it is crucial that the mass type (mono or avg) be set correctly, because the type of the mass calculated for the formula must be of the same type as the type of the data;
Increment charge by will iterate in all the items present in the list and apply the charge increment to them. For example, one item in the list that is charged 1 will be deionized and reionized to 2 (this calculation involves the ionization rule of the oligomer, and thus its ionization formula);
Reionization will iterate in all the items present in the list and apply the new ionization rule, defined in this groupbox widget.
Mass-based actions which involve processing the input m/z lists with numerical data representing masses:
Apply mass will iterate in all the items present in the list and apply the entered mass to them. The value entered by the user is a mass, not a m/z ratio. Thus, this computation involves, for each (m/z,z) pair in the list the sequential deionization, mass addition, reionization.
Apply threshold will remove all data items in the list for which m/z or M is less than the value set, depending on the radio button that is selected (On m/z value or On M value).
6.4.2 Output of the Calculations #
Simulations performed on a single input m/z list produce a m/z list that is identical to the input list, unless for the m and/or z values, which might have changed. This means that it is perfectly possible to:
Overwrite the initial data with the newly obtained ones (this is performed by checking the Perform computation in place check button widget);
Create a new list with the newly obtained data. As a convenience for the user, the new list will be an input m/z list in which it will be possible to perform ulterior simulations. This is useful when the simulations that need to be performed are sequential in kind. To have a new list created uncheck Perform computation in place.
6.4.3 Internal Workings #
When an operation is performed on the items of an input m/z list, say we want to make sodium adducts (that would be a formula “-H+Na”) of all the items in the list, the process involves the following steps, as detailed below for one single item of the list (which has data pair (334.341,3) and protonation as ionization agent).
Convert the tri-protonated analyte into a non-ionized analyte, thus getting M=1000;
Compute the mass of the “-H+Na” formula: 21.98 Da;
Add 1000+21.98;
Reionize to the initial charge state: (341.67,3).
6.5 Working on Two Input m/z Lists #
It is possible to perform calculations on two input m/z lists. These calculations are called matches. The (m/z,z) pairs of two different input m/z lists might be matched. Typically, a match operation would involve data from the mass spectrometer and data from a massXpert-based simulation (cleavage or fragmentation, for example). In order to perform a match operation, the first input m/z list (the data from the mass spectrometer) should be selected by its name in the Catalogue list and the second input m/z list (the data from the simulation) should be selected by its name in the Catalogue 2 list. Note that if the two input m/z lists are not of the same type (one is mono and the other is avg), the user will be alerted about this point.
6.5.1 Output of The Calculations #
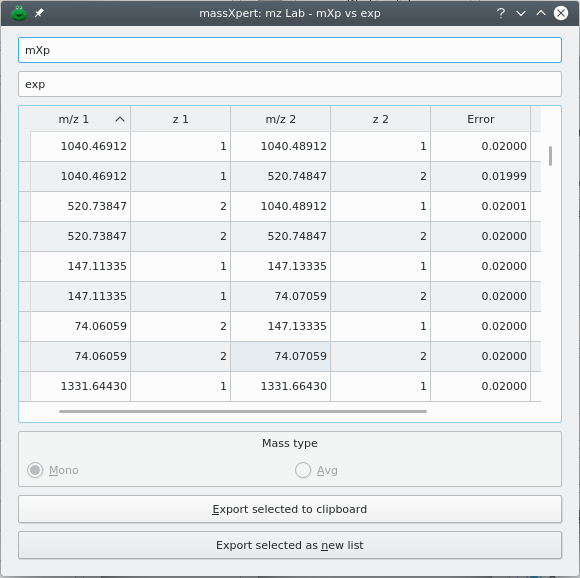
Calculations involving matches between two input lists produce an output that is displayed in an output m/z list, which is different from an input m/z list. Figure 6.6, “Match operation between two m/z lists, output list dialog window.” shows the results after having performed a match operation between an input m/z list obtained from the mass spectrometer (Catalogue 1) and an input m/z list obtained by simulating a cleavage with trypsin (Catalogue 2). The output m/z list dialog window holds all the matches along with the original data and the error.
Figure 6.6: Match operation between two m/z lists, output list dialog window. #
6.5.2 Tracing the Data #
When the data used for filling an input m/z list come from a massXpert-based simulation it is possible to trace back the (m/z,z) pair items to the corresponding sequence in the polymer sequence editor that gave rise to these oligomers in the first place. This is only possible if:
The way the data were fed into the input m/z list was by dragging oligomers from the treeview widgets, as described earlier;
The polymer sequence window is still opened when the tracing back is tried.
If the data do not originate immediately from the massXpert-based simulations (that is, the data do not originate directly from results treeview of cleavages or fragmentations or mass searches), it is still possible to perform the highlighting of corresponding oligomers in the sequence editor window, provided that the following requirements are met:
The data imported in the m/z list of m/z list are rich, that is, they comprise the coordinates data in the right format (see the data format, Section 6.3.1.4, “General Rules on Textual Mass Data Format”;
The proper polymer sequence is opened in a sequence editor window;
The identifier in the This window identifier line edit widget of the sequence editor window above is copied into the Sequence editor identifier line edit widget.
In the case the above conditions are met, double-clicking onto a item of the m/z list will highlight the corresponding sequence region in the sequence editor window.
In order to trace back any given item in an input or in an output m/z list to its corresponding polyemr sequence, just activate the item while having a look at the polymer sequence whence the oligomers initially originated. Each time an item is activated by double-clicking it, its corresponding sequence region will be highlighted (selected, actually) in the polymer sequence.